
Hello, I’m Salma.
AI Scientist
Data Science lead at the foundation model research team at SAP, building and fine-tuning large language models for enterprise applications.
Contact me!

Data Science lead at the foundation model research team at SAP, building and fine-tuning large language models for enterprise applications.
Contact me!

With an academic background in Machine Learning and Computational Biology, I currently work as a Senior AI scientist in the Foundation Model Research team at SAP. My work focuses on fine-tuning large language models and building Generative AI Applications. I currently lead the Data Science team at SAP, where we build AI assistants for ABAP developers.
My academic research focused on developing statistical and machine learning models (NLP) based on high-throughput sequencing datasets to better understand biological systems.
I like spending my free time hiking, reading popular science books, and playing board games. The pandemic encouraged me to rediscover my painting passion.
About MeLeading a data science team at SAP focusing on ABAP developer efficiency features for Joule (SAP's AI Copilot), including code completion, code generation, and code explanation.

SAP's premium Document Information Extraction service automates the processing of business documents like invoices and delivery notes with generative AI. This enables customizable extraction schemas and adds support for over 40 languages.
Open-source tool and web server for predicting spaced RNA sub-sequences attracting RNA-binding proteins.

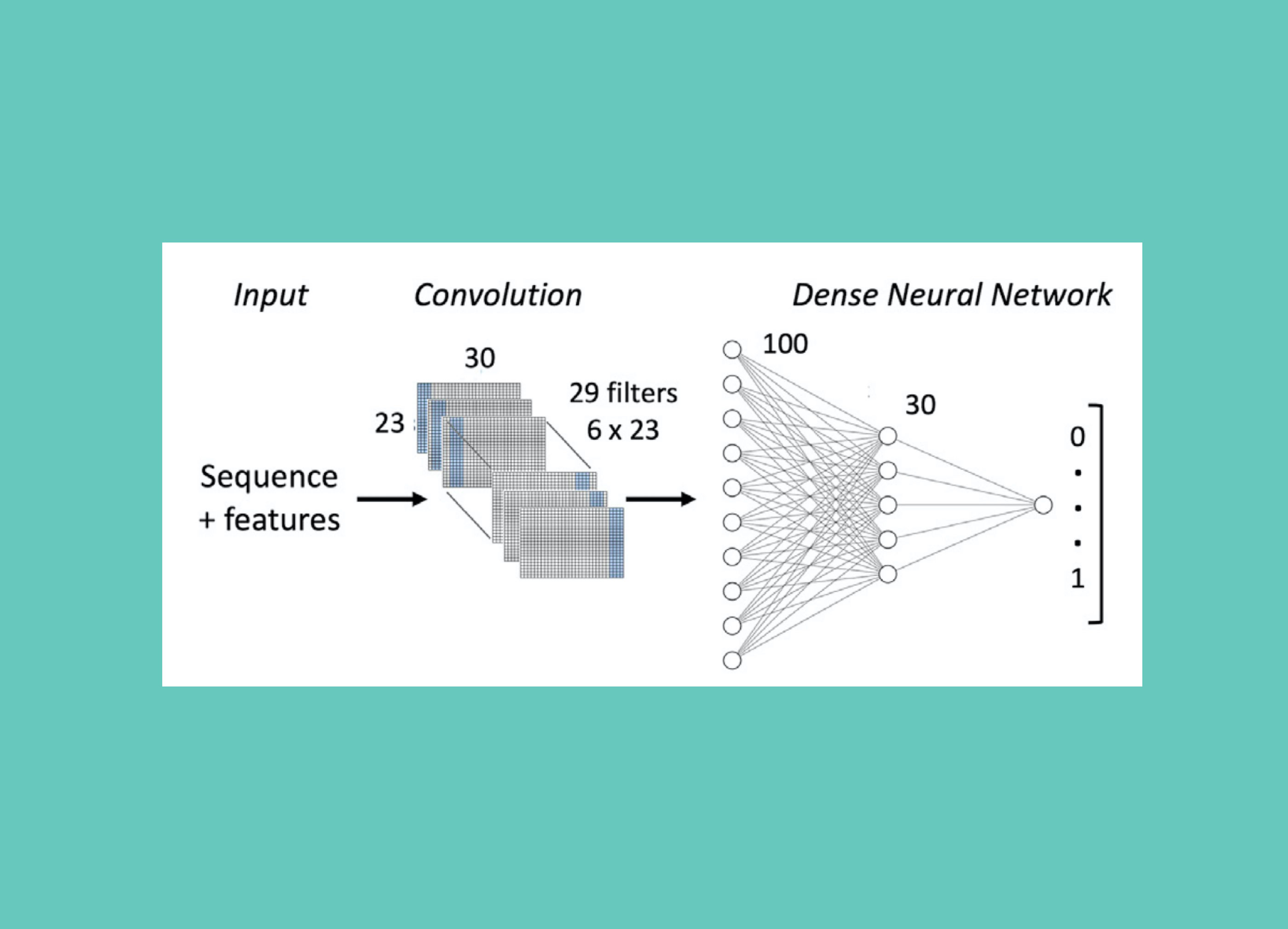
Leveraged CNNs to find protein sequences that can activate gene transcription.

Thanks for getting in touch!